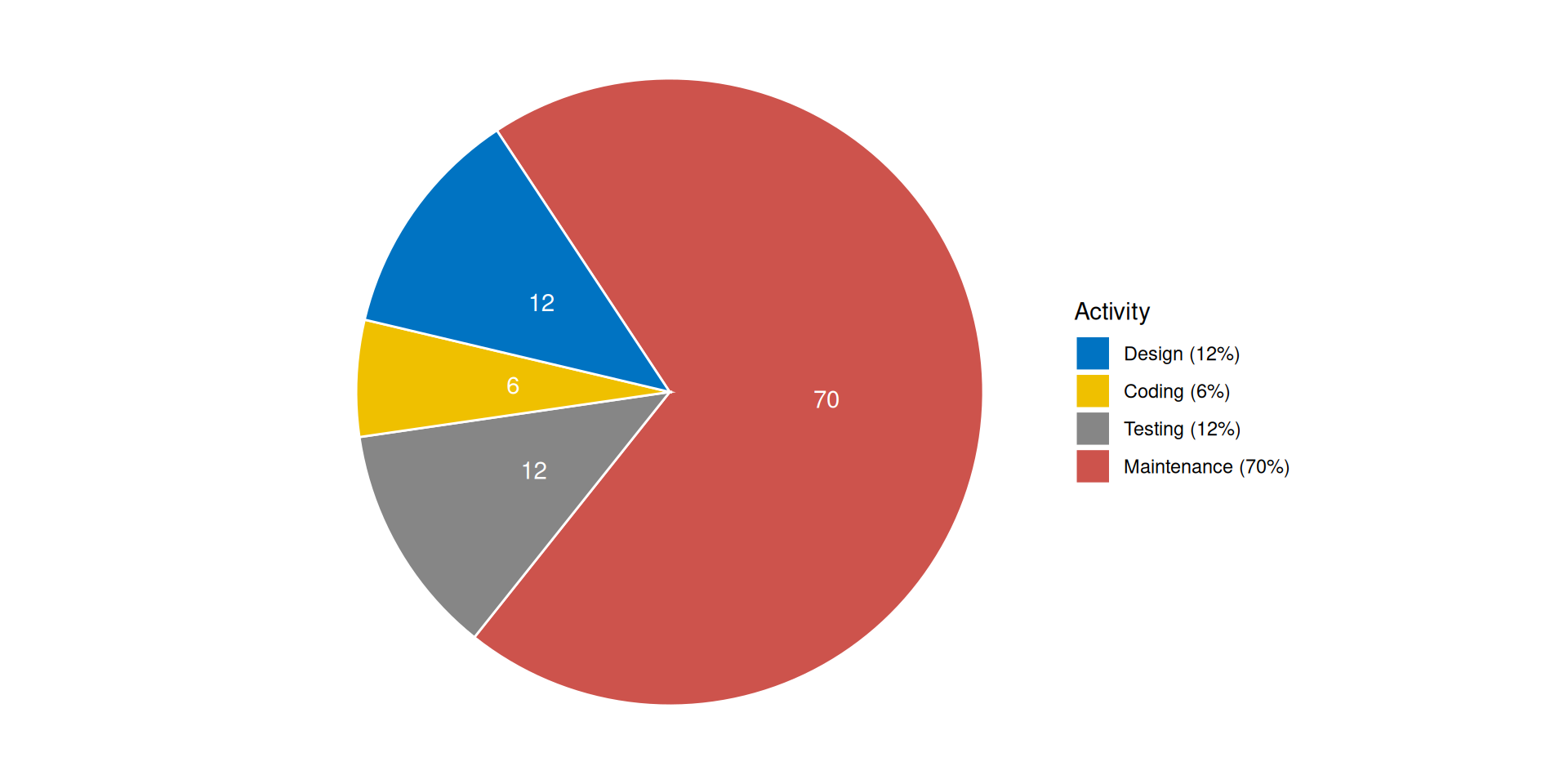
Cost distribution among software process activities
3 An R Package Engineering Workflow
Good Software Engineering Practice for R Packages
July 31, 2025
Motivation
From an idea to a production-grade R package
Example scenario: in your daily work, you notice that you need certain one-off scripts again and again.
The idea of creating an R package was born because you understood that “copy and paste” R scripts is inefficient and on top of that, you want to share your helpful R functions with colleagues and the world…
Professional Workflow

Photo CC0 by ELEVATE on pexels.com
Typical work steps
- Idea
- Concept creation
- Validation planning
- Specification:
- User Requirements Spec (URS),
- Functional Spec (FS), and
- Software Design Spec (SDS)
- R package programming
- Documented verification
- Completion of formal validation
- R package release
- Use in production
- Maintenance
Workflow in Practice

Photo CC0 by Chevanon Photography on pexels.com
Frequently Used Workflow in Practice
- Idea
- R package programming
- Use in production
- Bug fixing
- Use in production
- Bug fixing + Documentation
- Use in production
- Bug fixing + Further development
- Use in production
- Bug fixing + …
Bad practice!
Why?
Why practice good engineering?
Why practice good engineering?
Origin of errors in system development
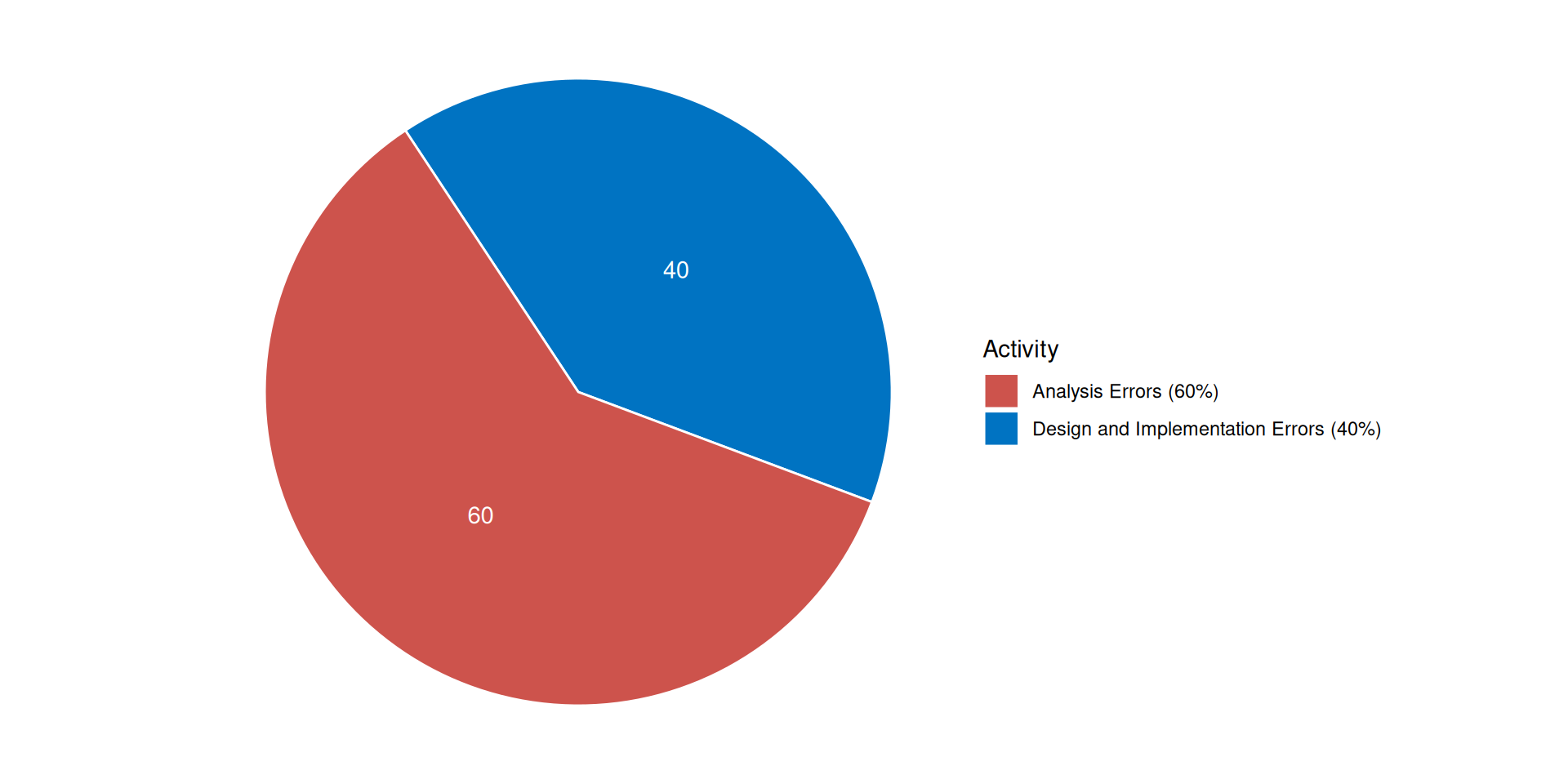
Boehm, B. (1981). Software Engineering Economics. Prentice Hall.
Why practice good engineering?
- Don’t waste time on maintenance
- Be faster with release on CRAN
- Don’t waste time with inefficient and buggy further development
- Fulfill regulatory requirements1
- Save refactoring time when the PoC becomes the release version
- You don’t have to be shy any longer about inviting other developers to contribute to the package on GitHub
Why practice good engineering?
Invest time in
- requirements analysis,
- software design, and
- architecture…
… but in many cases the workflow must be workable for a single developer or a small team.
Workable Workflow

Photo CC0 by Kateryna Babaieva on pexels.com
Suggestion for a Workable Workflow
- Idea
- Design docs
- R package programming
- Quality check (see Ensuring Quality by Chunyan)
- Use in production
Example - Step 1: Idea
Let’s assume that you used some lines of code to create simulated data in multiple projects:
Idea: put the code into a package
Example - Step 2: Design docs
- Describe the purpose and scope of the package
- Analyse and describe the requirements in clear and simple terms (“prose”)
| Obligation level | Key word1 | Description |
|---|---|---|
| Duty | shall | “must have” |
| Desire | should | “nice to have” |
| Intention | will | “optional” |
Example - Step 2: Design docs
Purpose and Scope
The R package simulatr shall enable the creation of reproducible fake data.
Package Requirements
simulatr shall provide a function to generate normal distributed random data for two independent groups. The function shall allow flexible definition of sample size per group, mean per group, standard deviation per group. The reproducibility of the simulated data shall be ensured via an optional seed It should be possible to print the function result. A graphical presentation of the simulated data will also be possible.
Example - Step 2: Design docs
Useful formats / tools for design docs:
- R Markdown1 (*.Rmd)
- Quarto1 (*.qmd)
- Overleaf2
- draw.io3
UML Diagram
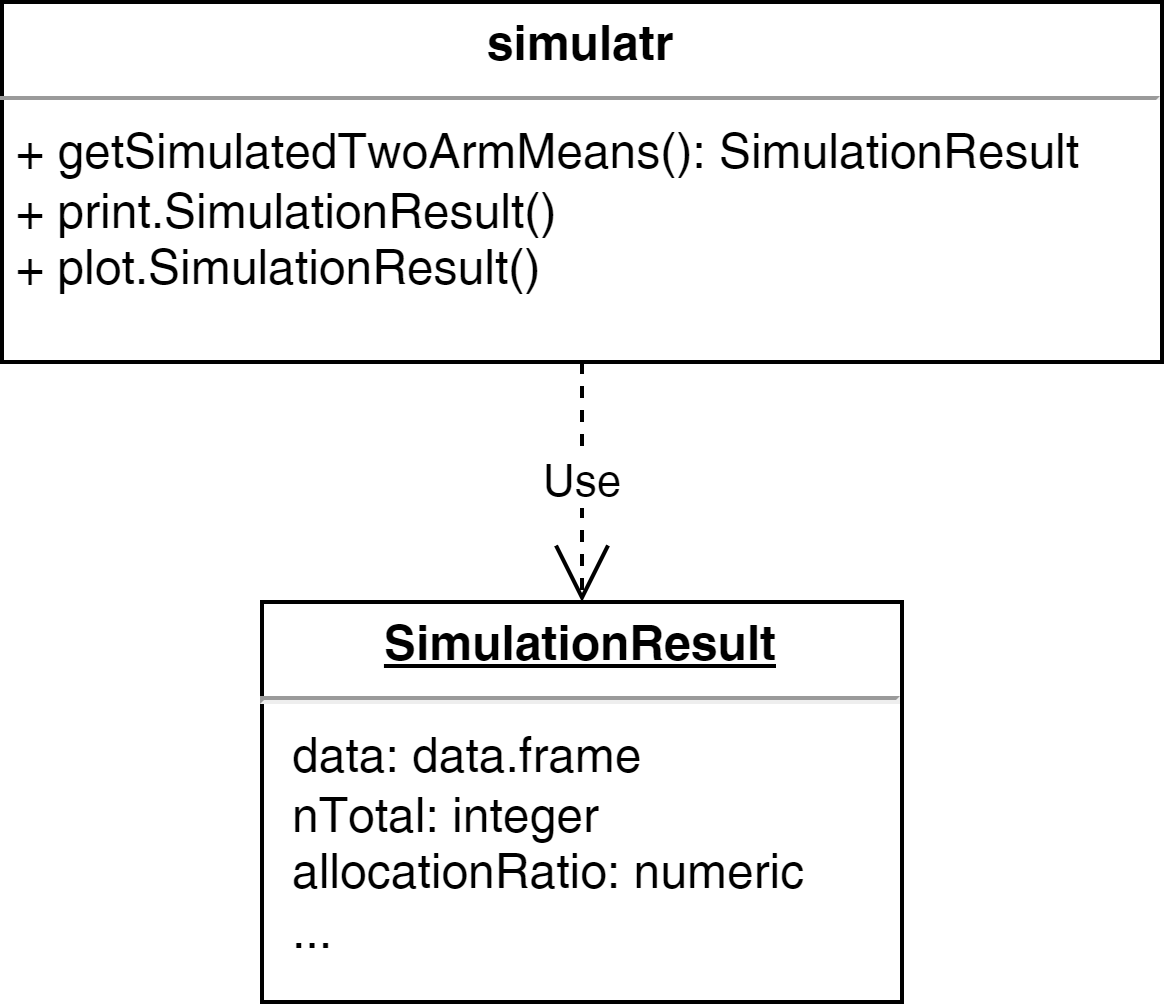
Example - Step 3: Packaging
R package programming
- Create basic package project (see R Packages by Shuang)
- C&P existing R scripts (one-off scripts, prototype functions) and refactor1 it if necessary
- Create R generic functions
- Document all functions
Example - Step 3: Packaging
One-off script as starting point:
Example - Step 3: Packaging
Refactored script:
Almost all functions, arguments, and objects should be self-explanatory due to their names.
Example - Step 3: Packaging
Define that the result is a list1 which is defined as class2:
getSimulatedTwoArmMeans <- function(n1, n2, mean1, mean2, sd1, sd2) {
result <- list(n1 = n1, n2 = n2,
mean1 = mean1, mean2 = mean2, sd1 = sd1, sd2 = sd2)
result$data <- data.frame(
group = c(rep(1, n1), rep(2, n2)),
values = c(
rnorm(n = n1, mean = mean1, sd = sd1),
rnorm(n = n2, mean = mean2, sd = sd2)
)
)
# set the class attribute
result <- structure(result, class = "SimulationResult")
return(result)
}Example - Step 3: Packaging
The output is impractical, e.g., we need to scroll down:
$n1
[1] 50
$n2
[1] 50
$mean1
[1] 5
$mean2
[1] 7
$sd1
[1] 3
$sd2
[1] 4
$data
group values
1 1 4.3085318
2 1 7.2507463
3 1 7.7603385
4 1 7.6181519
5 1 3.8011712
6 1 7.1346274
7 1 2.2246466
8 1 1.5797224
9 1 4.4450676
10 1 4.5412857
11 1 4.1412770
12 1 2.1803451
13 1 2.3093667
14 1 -0.3958166
15 1 2.6567318
16 1 10.3409040
17 1 8.1666885
18 1 7.9231871
19 1 4.6736150
20 1 4.2717743
21 1 3.3371229
22 1 5.8128029
23 1 9.0240783
24 1 2.3450643
25 1 -0.6780770
26 1 2.3545922
27 1 5.9192990
28 1 6.7119205
29 1 5.5885922
30 1 11.9070306
31 1 2.8536174
32 1 7.0711721
33 1 2.5451118
34 1 4.1594537
35 1 2.8693304
36 1 6.7134178
37 1 10.2538166
38 1 3.9364507
39 1 6.6141079
40 1 7.9941269
41 1 5.5201390
42 1 8.3276056
43 1 2.7852288
44 1 6.6187439
45 1 4.3804335
46 1 4.4176986
47 1 5.7012908
48 1 4.3768493
49 1 6.7289056
50 1 5.3188757
51 2 8.6597900
52 2 4.3406301
53 2 12.7796039
54 2 7.0045648
55 2 13.1952304
56 2 -2.1594604
57 2 11.0704184
58 2 5.8590565
59 2 4.5833703
60 2 2.3095432
61 2 4.9782035
62 2 15.3759790
63 2 9.5798835
64 2 5.5813900
65 2 14.1784838
66 2 9.8988104
67 2 6.2856296
68 2 11.8881048
69 2 6.2813503
70 2 1.4404367
71 2 6.1923174
72 2 8.8228714
73 2 4.3328003
74 2 7.2131012
75 2 0.8192615
76 2 7.1277341
77 2 13.0714286
78 2 10.2809193
79 2 10.3454593
80 2 6.2139426
81 2 6.3459276
82 2 14.5562710
83 2 5.8949658
84 2 10.5018679
85 2 6.6419736
86 2 9.2525678
87 2 12.8448539
88 2 2.4956992
89 2 2.2052089
90 2 7.1387222
91 2 3.9208955
92 2 5.8114928
93 2 1.6517912
94 2 9.2988980
95 2 7.2156933
96 2 14.1308951
97 2 5.5489472
98 2 6.6392445
99 2 0.7853674
100 2 13.1490879
attr(,"class")
[1] "SimulationResult"Solution: implement generic function print
Example - Step 3: Packaging
Generic function print:
#' @title
#' Print Simulation Result
#'
#' @description
#' Generic function to print a `SimulationResult` object.
#'
#' @param x a \code{SimulationResult} object to print.
#' @param ... further arguments passed to or from other methods.
#'
#' @examples
#' x <- getSimulatedTwoArmMeans(n1 = 50, n2 = 50, mean1 = 5,
#' mean2 = 7, sd1 = 3, sd2 = 4, seed = 123)
#' print(x)
#'
#' @export$args
n1 n2 mean1 mean2 sd1 sd2
"50" "50" "5" "7" "3" "4"
$data
# A tibble: 100 × 2
group values
<dbl> <dbl>
1 1 4.31
2 1 7.25
3 1 7.76
4 1 7.62
5 1 3.80
6 1 7.13
7 1 2.22
8 1 1.58
9 1 4.45
10 1 4.54
# ℹ 90 more rowsWebsite with pkgdown
Setup of pkgdown
pkgdownmakes it quick and easy to build a website for your package- After installing
pkgdown, just useusethis::use_pkgdown()to get started - Main configuration happens in
_pkgdown.ymlfile - Many customizations can be applied, but main work during development is to keep the
referencesection updated with names of.Rdfiles
Example _pkgdown.yml file
---
url: https://openpharma.github.io/mmrm
template:
bootstrap: 5
params:
ganalytics: UA-125641273-1
navbar:
right:
- icon: fa-github
href: https://github.com/openpharma/mmrm
reference:
- title: Package
contents:
- mmrm-package
- title: Functions
contents:
- mmrm
- fit_mmrm
- mmrm_control
- fit_single_optimizer
- refit_multiple_optimizers
- df_1d
- df_md
- componentPublication as GitHub Page
- It is helpful for users to read the website online
- GitHub is very helpful here because it allows
- A separate branch
gh-pagesthat stores the rendered website - GitHub actions automatically render the website when the
mainbranch is updated
- A separate branch
- To get started, use
usethis::use_pkgdown_github_pages()- Or, manually deploy site with
pkgdown::deploy_to_branch()
- Or, manually deploy site with
Exercise

Photo CC0 by Pixabay on pexels.com
Preparation
- Download the unfinished R package simulatr
- Extract the package zip file
- Open the project with RStudio
- Complete the tasks below
Tasks
Add assertions to improve the usability and user experience
Tip on assertions
Use the package checkmate to validate input arguments.
Example:
Error in playWithAssertions(-1) : Assertion on ‘n1’ failed: Element 1 is not >= 1.
Add three additional results:
- n total,
- creation time, and
- allocation ratio
Tip on creation time
Sys.time(), format(Sys.time(), '%B %d, %Y'), Sys.Date()
Add an additional result: t.test result
Add an optional alternative argument and pass it through t.test:
Implement the generic functions print and plot.
Tip on print
Use the plot example function from above and extend it.
Optional extra tasks:
Implement the generic functions
summaryandcatImplement the function
kableknown from the package knitr as generic. Tip: useto define kable as generic
Optional extra task1:
Document your functions with Roxygen2
- If you are already familiar with Roxygen2
References
- Gillespie, C., & Lovelace, R. (2017). Efficient R Programming: A Practical Guide to Smarter Programming. O’Reilly UK Ltd. [Book | Online]
- Grolemund, G. (2014). Hands-On Programming with R: Write Your Own Functions and Simulations (1. Aufl.).
O’Reilly and Associates. [Book | Online] - Rupp, C., & SOPHISTen, die. (2009). Requirements-Engineering und -Management: Professionelle, iterative Anforderungsanalyse für die Praxis (5. Ed.). Carl Hanser Verlag GmbH & Co. KG. [Book]
- Wickham, H. (2015). R Packages: Organize, Test, Document, and Share Your Code (1. Aufl.). O’Reilly and Associates. [Book | Online]
- Wickham, H. (2019). Advanced R, Second Edition.
Taylor & Francis Ltd. [Book | Online]
License information
- Creators (initial authors): Friedrich Pahlke
- In the current version, changes were done by (later authors): Joe Zhu
- This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
- The source files are hosted at github.com/pharmarug/pharmasug2025-r-workshop, which is forked from the original version at github.com/openpharma/workshop-r-swe.
- Important: To use this work you must provide the name of the creators (initial authors), a link to the material, a link to the license, and indicate if changes were made